What if machines learned how to generate novel, plausible, and testable research hypotheses from existing body of knowledge? That’s the entire promise behind Google DeepMind’s AI Co-Scientist, a multi-agent system that can look at existing literature and propose hypothesis that scientists can test and prove. In an era where there’s too much information, Google’s AI Co-Scientist points towards a future where AI works alongside scientists.
Built with Gemini 2.0, the AI Co-Scientist aims to solve the greatest bottleneck in hypothesis generation. Today’s researchers are often so fed up with too much information and an ever-expanding volume of literature. At the same time, making sense of that data and turning it into testable, impactful research questions is increasingly time-consuming, cognitively demanding, and limited by human bandwidth.
That’s where the AI Co-Scientist comes in. It’s a system that simulates the scientific process from hypothesis generation to peer review and experimental proposal using a coordinated team of AI agents. The Co-Scientist proposes a hypothesis and the scientist’s job is to verify the hypothesis and test its validity. In fact, independent researchers used the Co-Scientist’s generated hypothesis, tested it experimentally, and found good results.
This research holds transformative potential for a wide range of scientific fields. Chemists can use it to explore new reaction pathways or catalysts. Pharmaceutical companies can accelerate preclinical drug discovery. Environmental scientists can uncover overlooked patterns in ecological data. Whether you’re in academia or industry, the AI Co-Scientist represents a powerful shift in how scientific knowledge is created, tested, and applied.
How the AI Co-Scientist Works in General
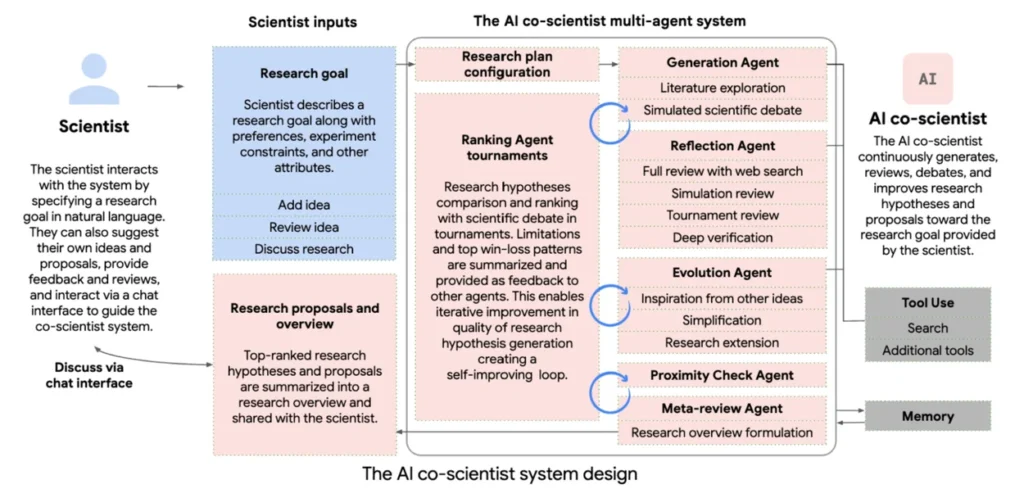
At its core, the AI Co-Scientist is designed to simulate the entire scientific method using a multi-agent large language model (LLM) system. Unlike single-output AI tools that just summarize papers or answer questions, this system mimics how scientists actually work: generating ideas, debating them, refining them, and proposing experiments, iteratively and autonomously.
The process starts with a research goal, to be written and explained by the scientist. This could be anything, from finding novel drugs for liver fibrosis to explaining the mechanisms behind antimicrobial resistance. Once done, the system kicks off a structured reasoning loop, breaking the problem down, parsing related literature, generating the hypothesis, and improving them over time through a series of debates and collaboration between the individual agents.
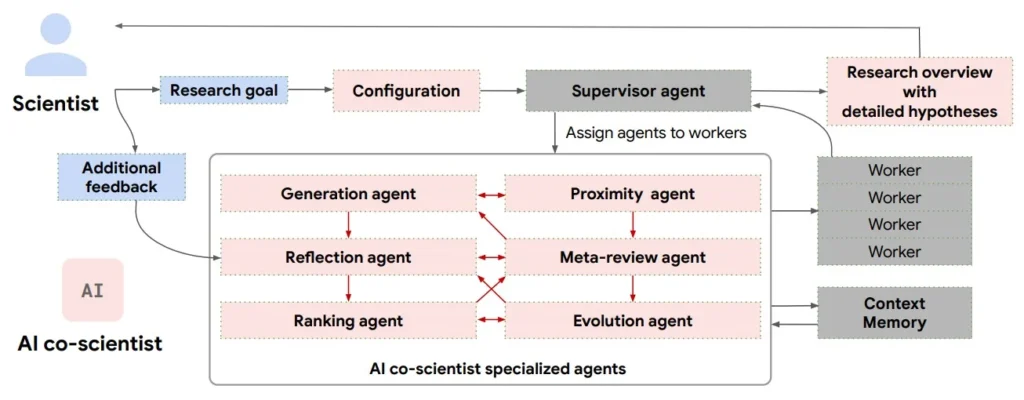
Each agent has a specific role, coordinated by a central Supervisor Agent, and all outputs are stored in a context memory that enables self-improvement across iterations. The process continues until a set of high-quality, testable hypotheses is produced. The AI Co-Scientist multi-agent system includes the following:
- Generation Agent: The starting point of the whole agent system. Its job is to interpret the research goal, search for related literature, and generate novel but plausible hypotheses, grounded in prior knowledge.
- Reflection Agent: This agent acts as a peer reviewer, performing a multi-stage critique of each hypothesis generated by the Generation Agent. It checks correctness, novelty, and feasibility, with or without access to external literature. It also decomposes hypotheses into underlying assumptions to test their soundness and even simulates biological or experimental outcomes step by step.
- Ranking Agent: This agent organizes the hypothesis into tournament-style matchups, testing them in pairwise debates. Using an Elo rating system, it scores each hypothesis based on how well it performs based on novelty, testability, and coherence.
- Proximity Agent: To avoid redundancy and promote diverse thinking, the proximity agents build a similarity graph across all hypotheses. It clusters similar ideas, flags duplicates, and helps identify underexplored regions in the research space. This informs which ideas to develop further or compare directly.
- Evolution Agent: This agent picks promising hypotheses and applies refinement strategies like improving grounding, simplifying language, combining concepts, or introducing alternative assumptions. Importantly, it doesn’t overwrite ideas; it generates new, evolved variants that re-enter the process.
- Meta-Review Agent: This agent steps back and looks at the whole system’s performance. It identifies patterns in critiques, surface recurring issues, and provides high-level summaries of the current research direction.
- Supervisor Agent (Coordinator): As mentioned, the supervisor agent handles the overall process – distributing tasks, monitoring progress, allocating resources, and triggering system resets based on convergence criteria or time budgets. It also manages the shared memory, which allows the agents to continuously learn from past iterations.
Together, these agents form a self-improving research loop. Ideas get generated, critiqued, ranked, refined, and re-tested until the system converges on a set of high-Elo, diverse, testable, and novel hypotheses.
Evaluating the Capability of the AI Co-Scientist
To assess whether the AI Co-Scientist produces high-quality scientific hypotheses, the researchers implemented a comprehensive evaluation framework. This included automated metrics, benchmark datasets, expert judgments, and comparisons with other language models and human researchers. The results show that the system not only generates plausible hypotheses, even improving it over time and outperforming strong baselines.
Elo Rating Correlates with Hypothesis Quality
The AI Co-Scientist used an Elo-style rating system (like Chess) to rank its generated hypotheses based on pairwise comparisons. To test whether this metric is meaningful, researchers evaluated the system’s Elo scores on GPQA, a benchmark of graduate-level science questions across biology, chemistry, and physics.
The findings showed a clear correlation: hypotheses with higher Elo scores were significantly more accurate and reliable. In fact, the AI’s top-ranked answers achieved 78.4% accuracy on GPQA, outperforming random selection and proving that Elo is a strong internal signal for hypothesis quality.
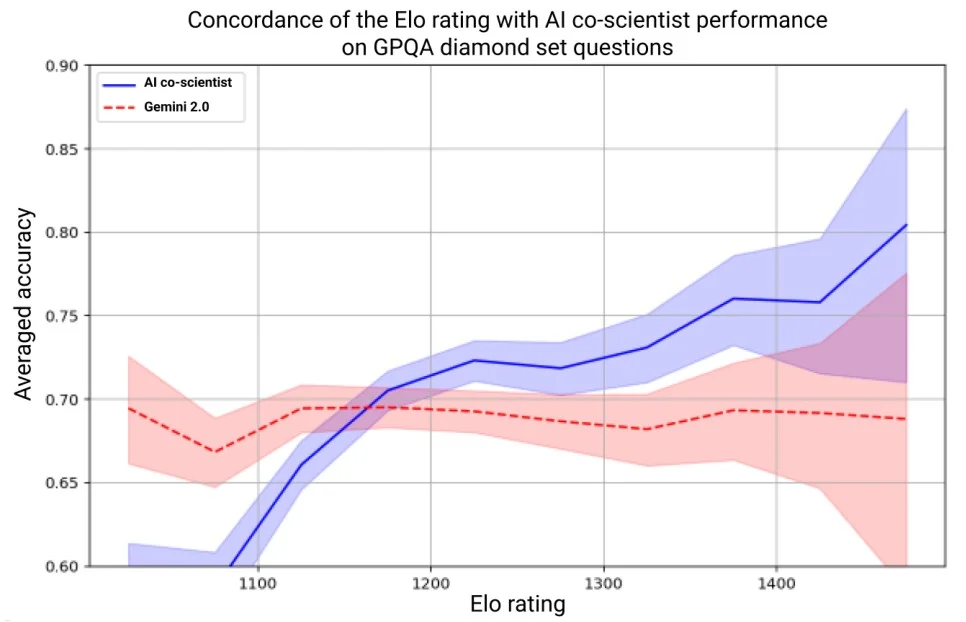
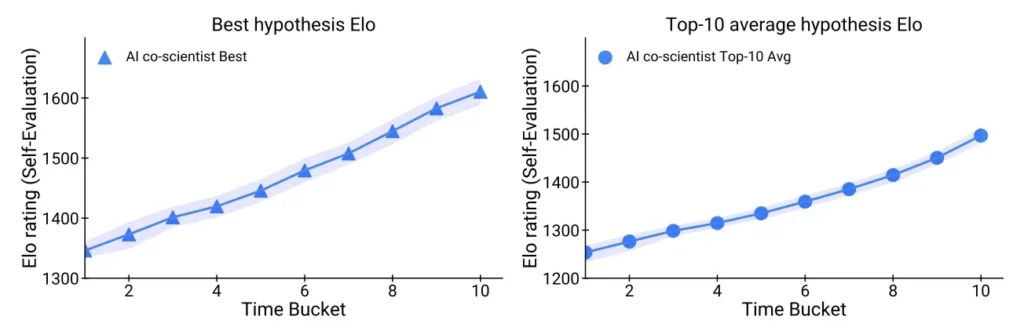
Test-Time Compute Improves Reasoning
Unlike static models, DeepMind researchers built the AI Co-Scientist to improve with time and through iterations. The researchers tested this by running the system longer across 203 research goals, allowing more rounds of generation, critique, ranking, and refinememt.
As compute increased, so did the performance. The hypotheses from later iterations consistently received higher Elo scores and improved in novelty and testability. This confirms that the system benefits from iterative self-improvement, making it more powerful the longer it runs.
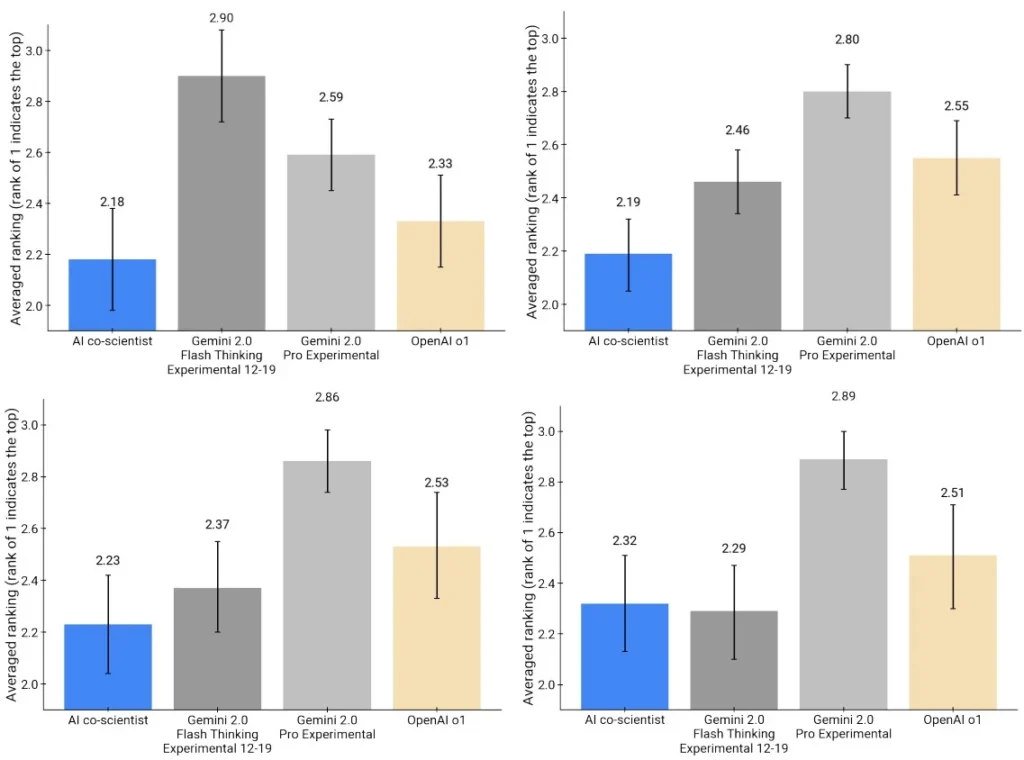
Comparison with Other LLMs and Experts
The AI Co-Scientist was also benchmarked against the following:
- Gemini 2.0 Pro Experimental
- OpenAI o1 and o3-mini-high
- DeepSeek R1
- Human experts tasked with writing their best hypothesis for the same biomedical problems.
On 15 expert-curated challenges, the AI Co-Scientist outperformed every other model and the human-written hypotheses, based on Elo ratings. Not only did it generate better ideas, but it also improved on the best expert suggestions after multiple iterations—highlighting the strength of its self-improving loop.
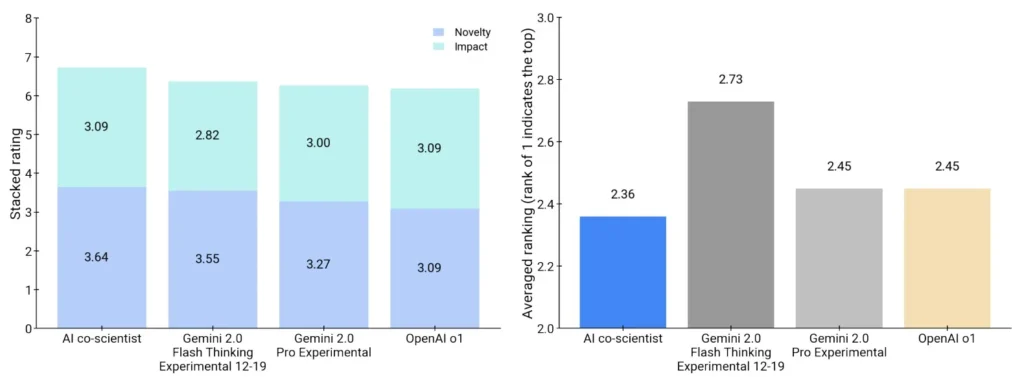
Expert Evaluation
Finally, the researchers brought in domain experts to judge the system’s outputs directly. Experts rated hypotheses from the Co-Scientist and three baseline LLMs across three criteria:
- Novelty – Is the idea original and previously unpublished?
- Impact – Could it lead to meaningful scientific advances?
- Overall Preference – Which output would they choose to pursue?
The Co-Scientist consistently scored higher in all categories:
- Novelty score: 3.64 out of 5
- Impact score: 3.09 out of 5
- Experts preferred Co-Scientist’s hypotheses over those from Gemini, OpenAI, and others.
Applying the AI Co-Scientist to Real-World Problems
To further prove the validity of the AI Co-Scientist, it was tested against real-world biomedical research. The researchers selected three major scientific challenges and used the system to generate hypotheses that were then passed on to human experts and validated in wet-lab experiments. The results demonstrate that the system is capable of contributing to real-world scientific discovery.
Drug Repurposing for Acute Myeloid Leukemia (AML)
In this task, the AI Co-Scientist was given a broad objective: identify new or overlooked drug candidates for treating AML, a complex and aggressive cancer. The system produced hypotheses that included both known and novel drug candidates:
- It correctly identified binimetinib and pacritinib, both of which are known to have AML-related activity.
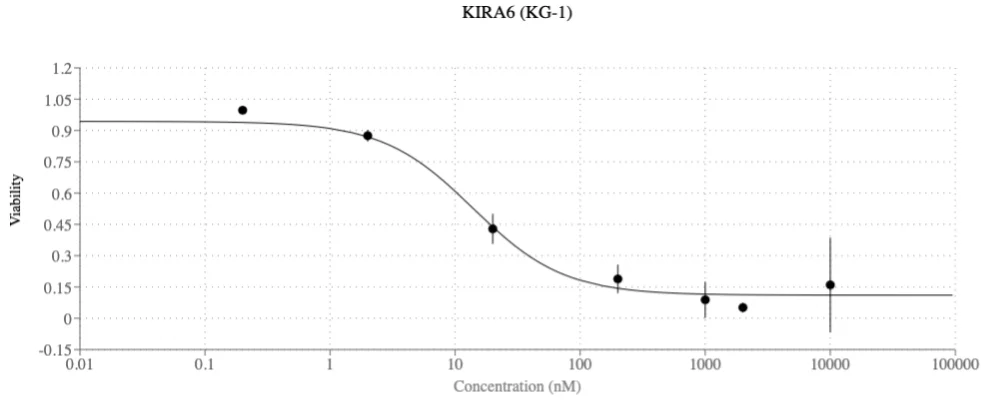
- More impressively, it proposed KIRA6, a drug not previously associated with AML treatment.
KIRA6 was taken into in vitro validation, where it showed nanomolar-level efficacy in inhibiting AML tumor cells. The AI identified KIRA6 in a hypothesis-driven context, and it performed successfully in lab testing.
Novel Targets for Liver Fibrosis
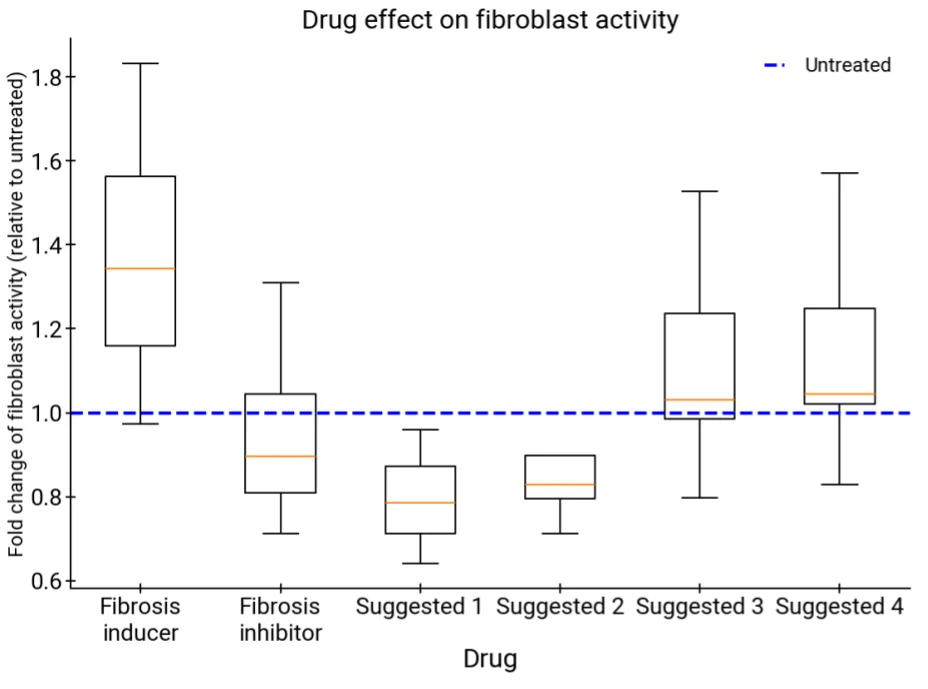
Liver fibrosis, a progressive and often irreversible scarring of liver tissue, currently lacks effective targeted therapies. The AI Co-Scientist was tasked with proposing new epigenetic targets that could reverse or slow the fibrotic process.
The system produced several hypotheses, three of which focused on understudied chromatin regulators. Two of the proposed targets were linked to available compounds that were then tested in human hepatic organoids (miniaturized models of human liver tissue).
The result showed that both compounds demonstrated clear anti-fibrotic effects, validating the AI’s ability to point toward therapeutically relevant molecular targets.
Mechanism of Antimicrobial Resistance
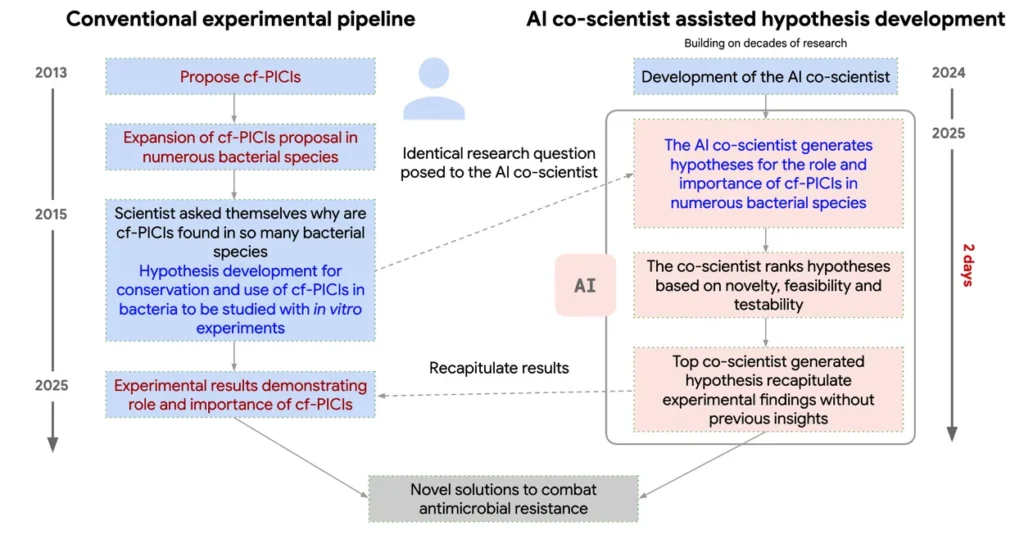
To push the limits of novelty, researchers posed a harder challenge involving an unpublished mechanism of bacterial gene transfer related to antimicrobial resistance, something known only to the human team, not present in the literature.
The AI Co-Scientist was tasked with explaining how antibiotic resistance could spread between bacterial species. After 2 days of iterative reasoning, the system produced a hypothesis remarkably similar to a recently discovered, unpublished mechanism involving cf-PICIs (phage-inducible chromosomal islands that carry resistance genes).
This result is particularly significant because it means the AI was able to infer a complex, multi-step biological mechanism that took humans a decade to uncover, using only open-access data and iterative reasoning.
Limitations & Challenges
While the AI Co-Scientist represents a major leap forward in using AI to accelerate scientific discovery, it’s not without limitations. Like any early-stage innovation, it comes with both technical and practical challenges that need to be addressed before widespread adoption is possible.
Limited Access to Scientific Literature
One of the most significant constraints is that the AI Co-Scientist is currently limited to open-access literature. That means it cannot access many of the most influential or cutting-edge papers locked behind paywalls. This poses a risk of missing critical prior work, generating redundant hypotheses, or overlooking important negative results that are often underreported in open literature.
Experimental, Not Yet Plug-and-Play
Although the system has demonstrated success in controlled biomedical settings, it’s still largely experimental. Deploying it in a general-purpose lab or research team would require:
- Careful prompt design,
- Custom agent configuration,
- Expert interpretation of outputs.
This isn’t yet a product researchers can use off-the-shelf. The system still depends on human review and intervention to validate outputs and guide its use responsibly.
No Specialized Equipment, But High Compute Costs
The AI Co-Scientist doesn’t need physical lab tools to run, but it does require significant computational resources, especially when running multiple iterations and agents in parallel.
While this is manageable for tech companies or research institutions, it could be a barrier for small academic labs, research teams in low-resource environments, and individual users without access to high-performance cloud computing.
As of now, cost and scalability remain obstacles to democratized access.
Evaluation Metrics Are Still Evolving
The system uses Elo scores based on simulated scientific debates to rank hypotheses. While this has shown a strong correlation with quality, it’s still a proxy, not a ground-truth measurement. There’s no universally accepted benchmark for hypothesis quality, and expert judgment, while helpful, introduces subjectivity.
Wider adoption will require more robust, transparent evaluation frameworks that can generalize across domains.
Generalization Outside Biomedicine Is Unproven
All current validations focus on biomedical research—drug discovery, disease mechanisms, and molecular targets. It remains unclear how well the system performs in other fields such as materials science, physics, and environmental science.
Transferring this architecture across disciplines will require domain-specific grounding, tailored databases, and new agent behaviors adapted to different types of data and experiments.
Risk of Overreliance or Misuse
As with all powerful AI systems, there’s a risk that users might over-trust the outputs or fail to critically evaluate them. Without proper guardrails, the system could propose:
- Biologically plausible but unsafe experiments,
- Ethically questionable ideas,
- Hypotheses that violate domain constraints if misused.
Wider adoption will depend not just on technical performance, but on responsible integration into human scientific workflows—with transparency, oversight, and accountability.
The Road Ahead
The biggest challenge before this becomes widely adopted is bridging the gap between lab performance and real-world usability. That means building user-friendly interfaces, reducing compute costs, integrating with real-world data systems, and developing field-specific adaptations. With continued development, the AI Co-Scientist could become a standard tool for hypothesis generation across disciplines, but it’s not quite there yet.
What do you think? Would you trust an AI system to help generate research hypotheses in your field? Would this change how you approach your next project? What would be its impact across the scientific community?
The possibilities are exciting but scary at the same time. But what we can do now is to embrace the fact that AI is here to stay. We’ve opened the Pandora’s Box, and our job is to ensure that tools like AI Co-Scientist remain ethical and humane.
Reference: This article is based on the findings and data presented in the original research study. For full details, methodologies, and supporting information, you may access the research here.