According to the World Health Organization, antimicrobial resistance (AMR) is one of the top global public health and development threats. It’s estimated that AMR is responsible for about 1.27 million global deaths in 2019. As such, it’s crucial to make rapid and cost-effective solutions to address this issue.
However, techniques like culture-based methods and polymerase chain reaction (PCR) techniques have a lot of drawbacks. For one, culture-based methods can take up to 96 hours to perform. These techniques also are very expensive and require a high level of expertise to ensure that AMR profiling is accurate and successful.
On the other hand, PCR methods are only limited to single targeted genes. This narrow focus means that PCR may not capture the full range of resistance mechanisms of microorganisms. In addition, the need for multiple gene assays to cover various resistance mechanisms contributes to the higher price range of PCR compared to culture-based methods.
In a recent study published in Nature’s Scientific Reports, researchers proposed using machine learning models in combination with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) to efficiently predict antibiotic resistance profiles for Staphylococcus epidermidis, a significant nosocomial pathogen.
The study used 303,195 microbial samples taken from the Database of Resistance against Antimicrobials with MALDI-TOF Mass Spectrometry (DRIAMS). This dataset involves over 4000 S. epidermidis samples per antibiotic for most drugs, collected from four different clinical sites across multiple years (2015—2018).
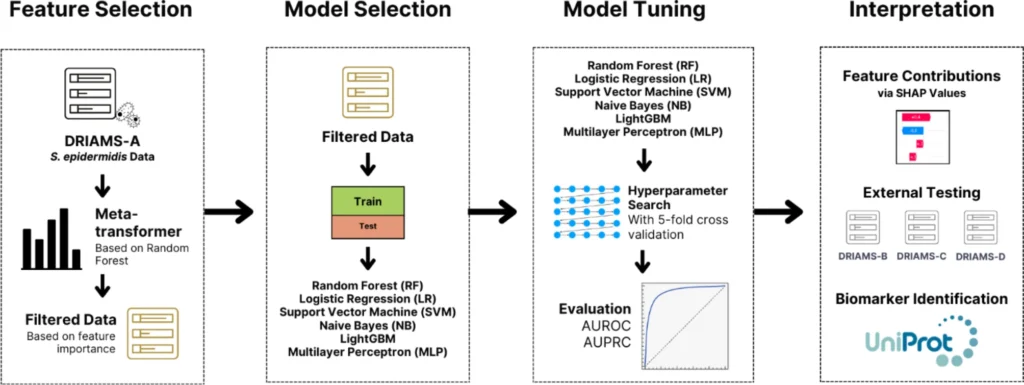
What’s interesting about this research is that they put more focus on data preprocessing and cleaning, which is always the best practice. They used the DRIAMS-A dataset, the largest collection in DRIAMS. DRIAMS B, C, and D were all then used for external validation.
Their data preprocessing workflow started with mass spectra preprocessing. Mass spectra often contain noise, baseline fluctuations, and irrelevant signals which need cleaning before training the model. Baseline removal and smoothing was done to eliminate any background noise and ensure that only meaningful spectral peaks remain for training.
Total ion current (TIC) normalization was also done to adjust intensity differences across spectra, which then prevents bias from varying sample concentrations. The researchers also trimmed to 2000-20,000 Da range to focus more on the most relevant protein signals in the spectra.
Lastly, binning was done (3 Da per bin) to reduce the complexity of spectral data. This converts raw spectra into structured numerical features that are easier for ML models to process.
After preprocessing, training the model started with feature selection using Random Forest. This method reduced the high-dimensional MALDI-TOF MS spectra from 6,000 to around 1,158 features per antibiotic.
For model selection, six machine learning algorithms were used, including LightGBM, Support Vector Machines (SVM), Random Forest, Logistic Regression, Naïve Bayes, and Multi-Layer Perceptron (MLP). After training and testing, LightGBM and SVM emerged as the top performing algorithms due to their ability to handle imbalanced and high-dimensional data.
Hyperparameter tuning was done using grid-search and cross-validation to optimize model performance. After training, the researchers interpreted the model using SHapley Additive exPlanations (SHAP) to identify the most influential features linked to antimicrobial resistance.
Finally, external validation on independent hospital datasets (DRIAMS B, C, and D) ensured generalizability and robustness across different clinical settings.
The study successfully applied ML algorithms to MALDI-TOF MS data to predict antibiotic resistance of S. epidermidis with a high accuracy (AUROC from 0.80-0.95 and an AUPRC of 0.97). The researchers also proved that the method they employed is much faster than traditional AST methods, reducing diagnostic times, from days to minutes, and only costing about $0.50 per test.
Using SHAP also allowed the researchers to identify key protein biomarkers linked to antibiotic resistance, including uncharacterized proteins that could be novel AMR markers. The researchers also noted that some identified features were associated with horizontal gene transfer and pathogenicity islands, which could provide insights into bacterial resistance evolution. This, however, was not elaborated further, and was left as an extension of the study.
It’s important to note that while the study focused on S. epidermidis, the workflow can be adapted for other bacteria such as Staphylococcus aureus, Escherichia coli, and Klebsiella pneumoniae. Doing so would make the algorithm a more versatile diagnostic tool for anti-microbial screening.
Let’s take a moment now to focus on why random forest was chosen as the algorithm for feature selection. Random forest is an ensemble learning method for classification and regression. The algorithm works by creating a multitude of decision trees during training and testing these trees for unknown and unseen data.
I believe that Random Forest (RF) was the best method for feature selection, in this context, because it’s capable of handling high-dimensional data, it ranks feature importance, and it prevents overfitting.
For instance, MS data generates thousands of spectral features, many of which are redundant and irrelevant. RF handles this large feature space without requiring explicit dimensionality reduction techniques like PCA. In addition, RF also assigns an Importance Score for each feature based on how much it improves the model accuracy. This allows the selection of only the most relevant spectral peaks associated with antibiotic resistance (~1,158 features per antibiotic).
Other methods like PCA, L1 or L2 regression, or Recursive Feature Elimination (RFE) could have been used but I think RF was preferred because it handles high-dimensional, imbalanced, and correlated data more efficiently. RF also maintains interpretability and biological relevance, making it the best fit for the study.
On the other hand, the study found that LightGBM and SVM worked well compared to the other classifiers used. I believe that’s because these algorithms are more suited for high-dimensional, structured data with complex patterns like MALDI-TOF MS data.
LightGBM can handle large feature spaces and imbalanced data efficiently using gradient boosting, making it more scalable and accurate than Random Forest. Also, SVM excels at detecting the most subtle patterns in complex datasets by using kernel functions, which help separate AMR-related features more effectively than linear models like Logistic Regression.
Despite being an artificial neural network algorithm, MLP didn’t work quite as well, probably because the MALDI-TOF MS data is high-dimensional but relatively small. When the dataset is small, deep learning techniques tend to overfit, which is why neural networks require thousands to millions of samples to generalize well.
Tree-based techniques like LightGBM performed better than MLP because they are more suited with structured and tabular data with limited samples. Additionally, MLP struggles with highly sparse feature spaces, while LightGBM and SVM handle these challenges more effectively.
Overall, the study proved how machine learning and AI can transform or enhance traditional antimicrobial resistance (AMR) detection by making it faster, more cost-effective, and scalable. For example, microbiologists and researchers studying antibiotic resistance mechanisms can utilize the method in this study for rapid AMR profiling and biomarker discovery.
Pharmaceutical and biotechnology companies can also use the method to identify resistance trends, guiding new antibiotic development. Those who specialize in clinical diagnosis can also integrate machine learning into their existing platforms.
Hospitals and clinics can also utilize this for rapid identification of drug-resistant infections to improve treatment decisions and reduce mortality rates, as highlighted by WHO. The method described in the study can also be integrated into antibiotic stewardship programs to optimize antibiotic use and slow the spread of antimicrobial resistance.
If ever someone wants to pursue this study further, some avenues for improvement could include:
- Prospective clinical trials: One of the questions I had in mind while reading the research was “How well does this method perform in real-time clinical settings?”. If ever, further studies can include testing the model on live samples instead of retrospective data. This can help in measuring live impact on clinical decision-making, patient recovery rates, and antibiotic stewardship.
- Model Adaptation in Different MALDI-TOF MS Machines: One can also explore the possibility of training models using data from different MALDI-TOF MS systems to improve the generalizability of the results.
- Handling Highly Imbalanced Datasets: Other techniques could also be used like SMOTE, cost-sensitive learning, or advanced deep learning methods to improve predictions for antibiotics with extreme resistance/susceptibility rates.
- Expansion to Other Bacterial Pathogens: You can also test the same approach on other clinically relevant bacteria such as Pseudomonas aeruginosa (resistant hospital infections), Klebsiella pneumoniae (carbapenem-resistant strains), or Escherichia coli (UTIs and bloodstream infections).
The fight against antibiotic resistance demands smarter, faster, and scalable solutions, and this study proved that machine learning is a tool we can use to revolutionize AMR detection. By combining machine learning techniques with MALDI-TOF MS, researchers unlocked a method that delivers accurate resistance profiling in minutes, at the fraction of the cost of traditional methods.
This finding not only enhances clinical diagnosis but also paves the way for AI-driven innovation in microbiology, pharmaceuticals, and global health surveillance.
However, technology won’t be enough. True impact comes from collaboration and real-world implementation. Researchers, clinicians, and data scientists must come together to refine, validate, and expand this approach, ensuring it becomes a standard tool against antimicrobial resistance.
Whether you’re a student, a researcher, or an industry professional, now is the time to act and explore how AI can transform chemistry and microbiology. Let’s build a smarter and better future – towards a data-driven global health.
Reference: This article is based on the findings and data presented in the original research study. For full details, methodologies, and supporting information, you may access the research here.